Smith-Root Staff
Thursday, November 19, 2020A new study, conducted by the Cawthron Institute, aimed to create a streamlined protocol for eDNA-based assessment of freshwater fish biodiversity in New Zealand. The report released this week details their systematic eDNA methods development process.
“The key to introducing a new protocol for monitoring New Zealand fish communities is identifying a method that, when compared with the standardized netting and electric fishing methods, is as effective, can be applied to a wide range of waterways, and is more economical.”
They compared multiple eDNA capture methods, two of which were selected for field testing and methodological comparison (Geotech pump with filtration manifold, vs. Smith-Root eDNA Sampler backpack and custom filter packets). “Within a sample reach, five methods of surface water sample collection were applied” – each a different combination of filter pore size and filtration method.

The researchers found that Geotech pump system was overall less efficient: “The Geotech pump could draw water through filters in the field using battery power but the time required to filter 1 L samples was extensive, i.e. greater than 60 min. There was also the potential for contamination with handling filters in the field.”
By contrast, the Smith-Root eDNA Sampler backpack and filters could filter multiple liters of water in a matter of minutes – maximizing the efficiency of valuable field technician time (See Table 1 from the report below). Smith-Root’s single-use eDNA filter packets also reduced potential risk of sample contamination in the field.
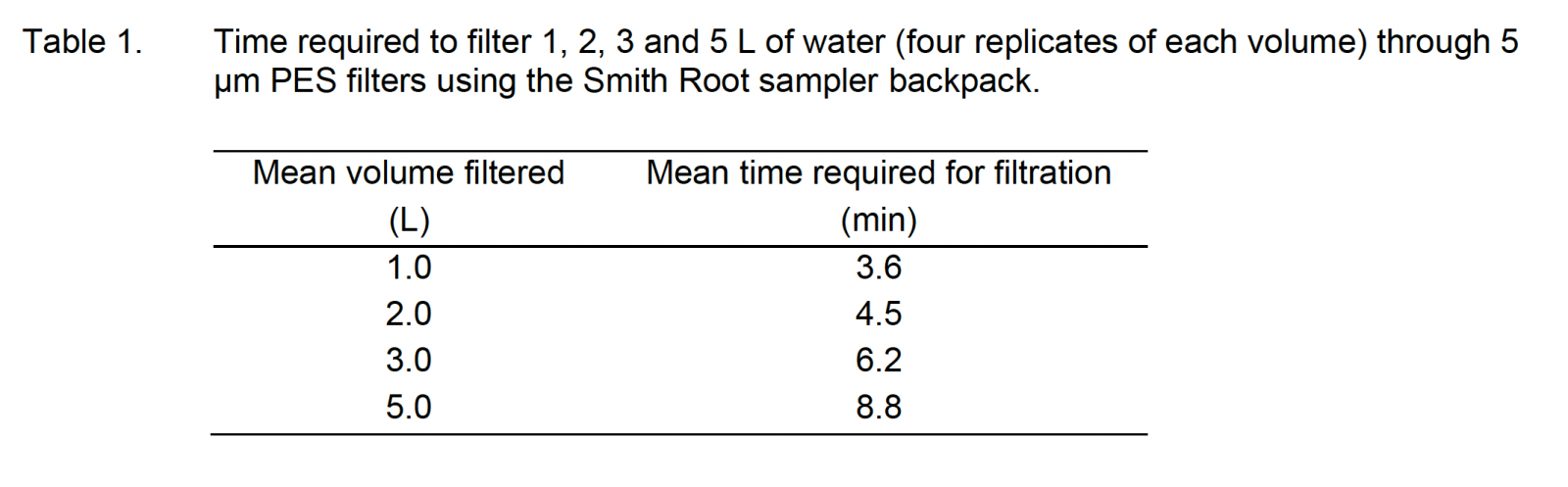
“The volume trial also revealed useful information regarding sample replication. Results suggest that to detect 100% of species present based on electric fishing records, more than three 3-L or 5-L samples are required.”
The study ultimately concluded that the Smith-Root sampling system outperforms other methods of eDNA capture.
“We found that a purpose-built backpack pump (eDNA sampler) facilitated the filtering of larger volumes of water (> 1 L) …compared with other methods tested (e.g. bench top, lab-based vacuum pumps) …and maximized fish eDNA detection”
The New Zealand project was also one of the first studies to evaluate the new self-preserving eDNA filters built by Smith-Root. The authors recommended self-preserving filters for future eDNA projects.
“Self-preserving filters are also available (and were used in later field trials), whereby there is no need to remove the filter from the housing (minimizing risk of contamination) and samples can be stored at room temperature prior to analysis”.
The study goes on to establish protocols for eDNA extraction from filter samples, PCR amplification/sequencing of fish eDNA, and bioinformatic analysis of sequence data to determine fish community composition.
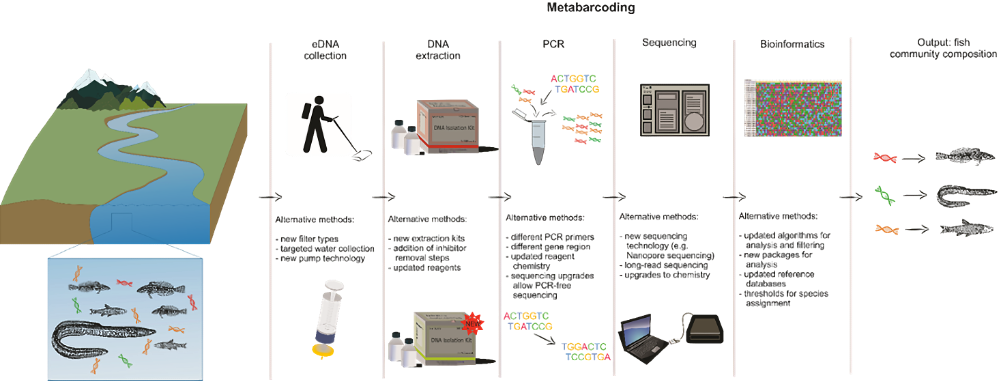
“Each step of the workflow can be considered somewhat modular; that is, the protocols can be updated or exchanged as technology changes and matures, although, as with sampling protocol changes using other technologies, these should be validated by running them alongside the existing protocols to ensure the outcomes are comparable.”